Metaphorically speaking, every human cell contains in its nucleus a huge library of tens of thousands of books, the genes. Each of these books in turn contains the building instructions for a protein. When the cell needs a certain protein, a transcription of the corresponding instructions is made. These transcriptions are called mRNAs (RNA is a slightly modified form of DNA).
A cellular mechanism ensures that the mRNA transcriptions are "shredded" again after a short time. This ensures that the protein is only produced as long as it is actually needed. Several decades ago, researchers came up with the idea of using this shredder for their own purposes: By specifically attaching a marker to certain mRNAs, they ensure that the transcriptions are not used as building instructions at all, but are destroyed immediately: a process also known as RNA silencing. The cell then lacks the corresponding protein. This makes it possible to find out which function it would actually be responsible for.
Bacterial molecule as light-dependent switch
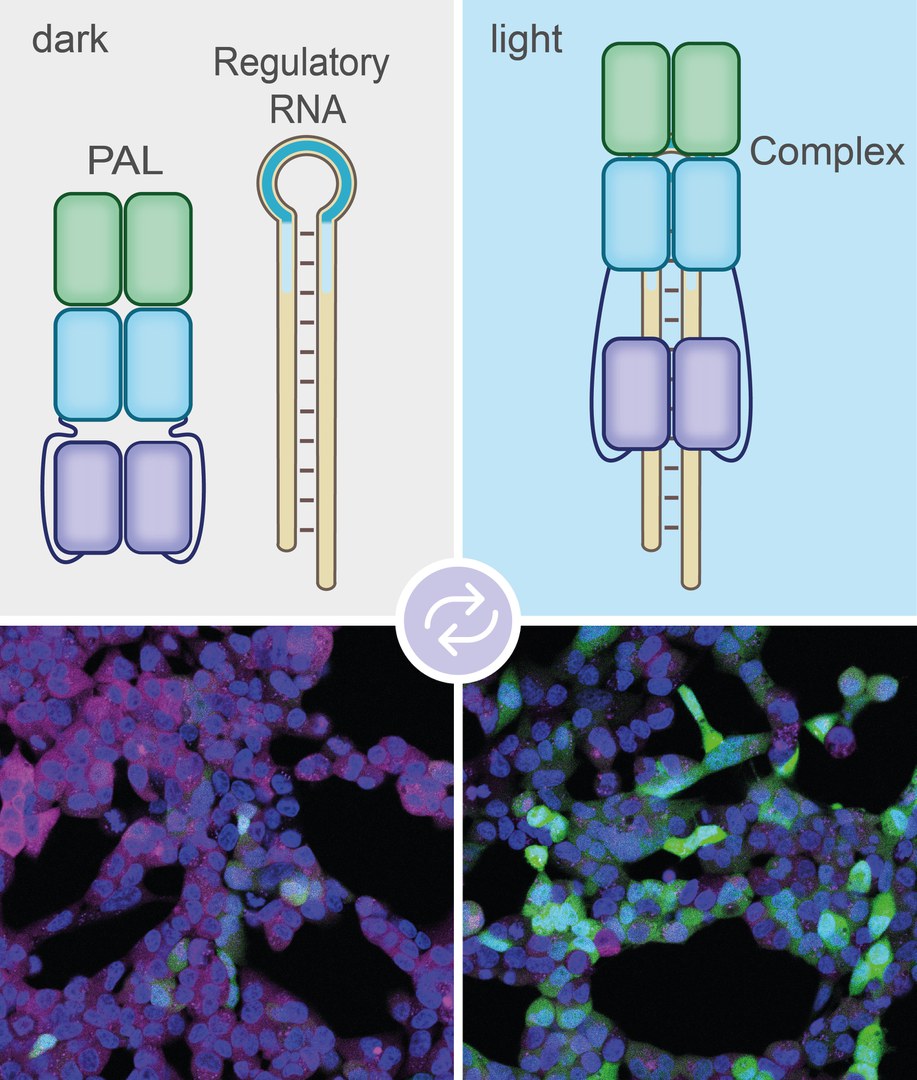
The approach that the groups from Bonn and Bayreuth have now published is based on this method. However, it is nowhere near as crude, but allows a far more differentiated control over the lifespan of the mRNA copies. "We use a bacterial molecule to control the shredding of mRNA transcriptions with the help of light," explains Prof. Dr. Günter Mayer, who heads the Chemical Biology & Medicinal Chemistry Research Group at the LIMES Institute of the University of Bonn.
The bacterial molecule with the abbreviation PAL acts as a kind of switch. It changes its shape under the influence of blue light. In the process, a pocket is exposed that can bind to certain molecules. "We searched a huge library of artificially produced short RNA molecules called aptamers," says Mayer. "Eventually we came across an aptamer that's a good match for the pocket in the PAL molecule."
The researchers have now coupled this aptamer to one of the molecular markers that can attach to mRNAs and thereby release them for degradation. "When we irradiate the cell with blue light, PAL binds to the marker via the aptamer and thus puts it out of action," explains Mayer's colleague Sebastian Pilsl. "The mRNA is then not destroyed, but translated into the corresponding protein." As soon as the researchers switch off the blue light, PAL releases the label again. Now it can attach itself to the mRNA, which is then shredded.
This will in future enable researchers to investigate exactly where and when a protein is needed in a cell, simply by immersing an area of the cell in blue light at a certain time and then looking at the consequences. In the current study they applied this to proteins that play an important role in the regulation of the cell cycle and cell division. The combination of aptamer and degradation marker is introduced into the cell by genetic engineering. This means that it generates the light-dependent degradation signal itself; it does not have to be supplied from outside.
Gene transcriptions can be specifically switched off
The aptamer can be combined with any markers, each of which in turn serves as a shredder signal for a specific mRNA. "This method can therefore be used to switch off practically every mRNA molecule in the cell in a controlled manner," emphasizes Prof. Dr. Andreas Möglich from the University of Bayreuth. In the recently published pilot study, it all worked both simply and reliably. The researchers therefore see great potential in their method for the investigation of dynamic processes in living cells and organisms.
Publication: Sebastian Pilsl, Charles Morgan, Moujab Choukeife, Andreas Möglich and Günter Mayer: Optoribogenetic control of regulatory RNA molecules; Nature Communications; dx.doi.org/10.1038/s41467-020-18673-5
Contact:
Prof. Dr. Günter Mayer
LIMES-Institut der Universität Bonn
Tel. +49 (0)228/73-2935
E-mail: gmayer@uni-bonn.de